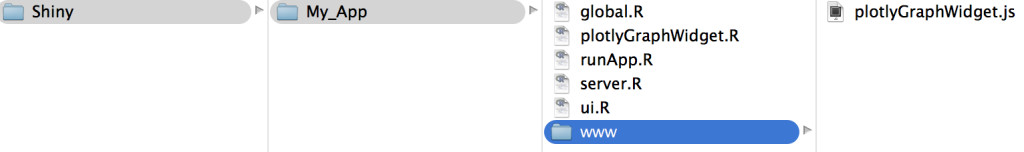
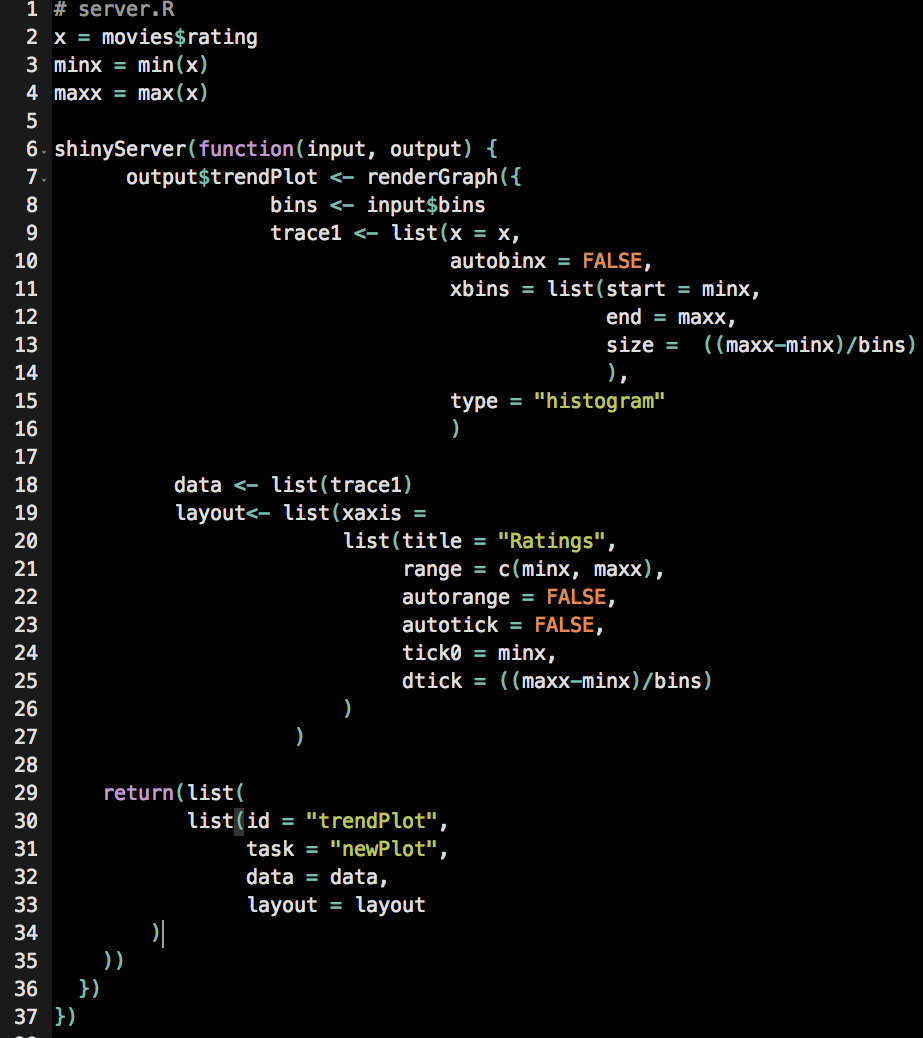
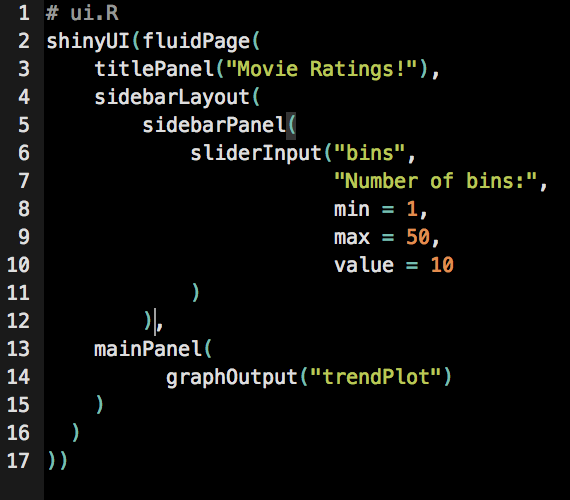
Function definition
# Function Definition ---------------------------------------------------------
VoronoiPlotly <- function(fit, # Fit object from K-Means
ds, # Data frame containing original data and clusters from K-Means
n.sd.x = 3, # Controls the width of the plot
n.sd.y = 3, # Controls the height of the plot
print.ggplot = FALSE, # Plots a diagnostic chart using ggplot2
point.opacity = 0.8,
point.size = 7,
point.symbol = "circle",
point.linewidth = 2,
point.lineopacity = 0.5,
plot_bgcolor = "#ffffff",
paper_bgcolor = "#ffffff",
center.size = 15,
shapes.opacity = 0.5,
shapes.linecolor = "#404040",
center.color = "#000000"){
# Options
options(stringsAsFactors = F)
graphics.off()
# Load libraries ------------------------------------------------------------
library(plotly)
library(deldir)
# Create convenience data frames ----------------------------------------------
centers <- data.frame(fit$centers)
vor <- deldir(centers)
# Calculate slopes
vor.df <- data.frame(vor$dirsgs, m = (vor$dirsgs$y2- vor$dirsgs$y1)/(vor$dirsgs$x2 - vor$dirsgs$x1))
# Calculate constants
vor.df$c <- with(vor.df, ((y2 - m*x2) + (y1 - m*x1))/2)
# Covnert to strings for better matching later on
vor.df.str <- data.frame(C1 = apply(vor.df[,1:2], 1, paste, collapse = ","),
C2 = apply(vor.df[,3:4], 1, paste, collapse = ","))
# Combine the x and y coordinates for each segment
coord.df <- rbind(as.matrix(vor.df[1:2]), as.matrix(vor.df[,3:4]))
# Convert to string
coord.df.str <- apply(coord.df, 1, paste, collapse = ",")
# Find unique strings
count <- sapply(coord.df.str, function(x){sum(coord.df.str == x)})
coord.df.str <- data.frame(str = coord.df.str, count = count)
coord.df.str <- subset(coord.df.str, count == 1)
# Get outer boundary co-ordinates
outer.bound <- matrix(as.numeric(unlist(strsplit(coord.df.str$str, ","))), ncol = 2, byrow = T)
outer.bound <- data.frame(x = outer.bound[,1], y = outer.bound[,2])
# Add respective slopes and constants
for(i in 1:nrow(outer.bound)){
str <- coord.df.str[i,1]
idx <- ifelse(is.na(match(str, vor.df.str$C1)), match(str, vor.df.str$C2), match(str, vor.df.str$C1))
# Slope
outer.bound$m[i] <- vor.df$m[idx]
# Constants
outer.bound$c[i] <- vor.df$c[idx]
}
# Find enclosing rectangle boundaries -----------------------------------------
x.min <- mean(ds$x) - n.sd.x*sd(ds$x)
x.max <- mean(ds$x) + n.sd.x*sd(ds$x)
y.min <- mean(ds$y) - n.sd.y*sd(ds$y)
y.max <- mean(ds$y) + n.sd.y*sd(ds$y)
# Create x-axsi and y-axis limits
xlim <- c(x.min, x.max)
ylim <- c(y.min, y.max)
# Extend outer boundary points to above rectangle ------------------------------
for(i in 1:nrow(outer.bound)){
# Extract x-y coordinates
x <- outer.bound$x[i]
y <- outer.bound$y[i]
# Get slope
m <- outer.bound$m[i]
# Get slope
c <- outer.bound$c[i]
# Extend to each edge of enclosing rectangle
ext.coord <- mat.or.vec(4,3)
# Extend to left edge
ext.coord[1,1] <- x.min
ext.coord[1,2] <- m*x.min + c
ext.coord[1,3] <- sqrt((ext.coord[1,1] - x)^2 + (ext.coord[1,2] - y)^2)
# Extend to right edge
ext.coord[2,1] <- x.max
ext.coord[2,2] <- m*x.max + c
ext.coord[2,3] <- sqrt((ext.coord[2,1] - x)^2 + (ext.coord[2,2] - y)^2)
# Extend to top edge
ext.coord[3,2] <- y.max
ext.coord[3,1] <- (y.max - c)/m
ext.coord[3,3] <- sqrt((ext.coord[3,1] - x)^2 + (ext.coord[3,2] - y)^2)
# Extend to bottom edge
ext.coord[4,2] <- y.min
ext.coord[4,1] <- (y.min - c)/m
ext.coord[4,3] <- sqrt((ext.coord[4,1] - x)^2 + (ext.coord[4,2] - y)^2)
# Find the closest edge
idx <- which.min(ext.coord[,3])
x <- ext.coord[idx,1]
y <- ext.coord[idx,2]
# Insert into outer bound
outer.bound$x.ext[i] <- x
outer.bound$y.ext[i] <- y
}
# Convert to string for easier searcing later on
outer.bound.str <- apply(outer.bound[,5:6], 1, paste, collapse = ",")
# Augment vor.df with extended outer bound coordinates -------------------------
for(i in 1:nrow(outer.bound)){
# Convert to string to help matching
str <- paste(outer.bound[i,1:2], collapse = ",")
# Match with original vor.df
if(is.na(match(str, vor.df.str$C1))){
idx <- match(str, vor.df.str$C2)
vor.df[idx, 3:4] <- outer.bound[i, 5:6]
}else{
idx <- match(str, vor.df.str$C1)
vor.df[idx, 1:2] <- outer.bound[i, 5:6]
}
}
# Plot Check ------------------------------------------------------------------
p.ggplot <- ggplot() +
geom_point(data = centers, aes(x, y), color= "red", size = 5) +
geom_point(data = ds, aes(x, y, color = cluster)) +
geom_segment(data = vor.df, aes(x = x1, y = y1, xend = x2, yend = y2)) +
geom_point(data = as.data.frame(fit$centers), aes(x, y)) +
geom_text(data = centers, aes(x,y, label = 1:nrow(centers)), size = 10) +
geom_point(data = outer.bound, aes(x.ext, y.ext), color = "blue", size = 5) +
geom_point(data = outer.bound, aes(x, y), color = "red", size = 5) +
geom_hline(yintercept = y.min) +
geom_hline(yintercept = y.max) +
geom_vline(xintercept = x.min) +
geom_vline(xintercept = x.max)
p.ggplot <- ggplotly(p.ggplot)
if(print.ggplot == T){print(p.ggplot)}
# -----------------------------------------------------------------------------
# Function to calculate which side of line is point on ------------------------
sideFUNC <- function(x, y, x1, y1, x2, y2){
d <- (x - x1)*(y2-y1) - (y - y1)*(x2 - x1)
return(round(d,2))
}
# Figure out the path for each polygon ----------------------------------------
path <- list()
# Loop thorough each centroid and find corrosponding edges
for(i in 1:nrow(centers)){
# Find each row where centeroid is available
mat <- subset(vor.df, ind1 == i | ind2 == i)
# Find all unique coordinates associated with centroid
mat <- cbind(matrix(c(mat$x1, mat$x2), ncol = 1), matrix(c(mat$y1, mat$y2), ncol = 1))
mat <- unique(mat)
mat.str <- apply(mat, 1, paste, collapse = ",")
# print(mat)
# Find all outer boundary points asociated with centroid
# If an outer boundary point is found, there must be atleast two
idx <- outer.bound.str %in% mat.str
if(sum(idx) == 2){
# Only if two outer boundary points are found
# then need to modify matrix and add edge end points
# Find the side where all other outer boundary points are
# Assuming all other boundary points are on the same side
# need only one point to find this out
p <- as.numeric(unlist(strsplit(outer.bound.str[!idx][1], split = ",")))
# Line segment is defined by the two identified outer boundary points
p1 <- as.numeric(unlist(strsplit(outer.bound.str[idx][1], split = ",")))
p2 <- as.numeric(unlist(strsplit(outer.bound.str[idx][2], split = ",")))
# Find side
side <- sideFUNC(p[1], p[2], p1[1], p1[2], p2[1], p2[2])
# Case when only two cluster and hence only one dividing segment
if(is.na(side)){
side <- sideFUNC(centers[i,1], centers[i,2], p1[1], p1[2], p2[1], p2[2])
}
if(side != 0){
# Find the enclosing rectangle"s endpoints that are on the opposite side
# Top - Left
side.check <- sideFUNC(x.min, y.max, p1[1], p1[2], p2[1], p2[2])
if(side.check != 0){if(sign(side.check) != sign(side)) {mat <- rbind(mat, c(x.min, y.max))}}
# Bottom - Left
side.check <- sideFUNC(x.min, y.min, p1[1], p1[2], p2[1], p2[2])
if(side.check != 0){if(sign(side.check) != sign(side)) {mat <- rbind(mat, c(x.min, y.min))}}
# Top - Right
side.check <- sideFUNC(x.max, y.max, p1[1], p1[2], p2[1], p2[2])
if(side.check != 0){if(sign(side.check) != sign(side)) {mat <- rbind(mat, c(x.max, y.max))}}
# Bottom - Right
side.check <- sideFUNC(x.max, y.min, p1[1], p1[2], p2[1], p2[2])
if(side.check != 0){if(sign(side.check) != sign(side)) {mat <- rbind(mat, c(x.max, y.min))}}
}
}
# print(mat)
# readline("Enter:")
# Re-order the points to ensure it makes a convex polygon
mat <- mat[chull(mat),]
#Paste together
path[[i]] <- paste0("M", paste0(mat[1,], collapse = ","))
path[[i]] <- paste(path[[i]],
paste(apply(matrix(mat[-1,], ncol = 2), 1, function(x){
vec <- paste0(x, collapse = ",")
vec <- paste0("L", vec)
}), collapse = " "),
"Z")
}
# Finally plot using Plotly ---------------------------------------------------
# crate a "shapes" list for voronoi polygons to be passed to layout()
shapes <- list()
cols <- RColorBrewer::brewer.pal(nrow(centers), "Paired")
# Loop through each path and add params like fill color, opacity etc
for(i in 1:length(path)){
shapes[[i]] <- list(type = "path",
path = path[[i]],
fillcolor = cols[i],
opacity = shapes.opacity,
line = list(color = shapes.linecolor))
}
# Change colors for each cluster to allow manual spec
for(i in 1:nrow(centers)){
ds$color[ds$cluster == i] <- cols[i]
}
# Create plot
# base layer
p <- plot_ly(ds, x = x, y = y , mode = "markers", name = "Clusters", opacity = point.opacity,
hoverinfo = "x+y+text",
text = paste("Cluster:",cluster),
marker = list(symbol = point.symbol, color = color, size = point.size,
line = list(color = "#262626", width = point.linewidth, opacity = point.lineopacity)),
showlegend = F)
# Add centroids
p <- add_trace(centers, x = x, y = y, mode = "markers", name = "Cluster Centers",
hoverinfo = "none",
marker = list(color = center.color, symbol = "cross", size = center.size))
# Add polygons
p <- layout(title = "Voronoi polygons and K- Means clustering",
paper_bgcolor = paper_bgcolor,
plot_bgcolor = plot_bgcolor,
xaxis = list(range = xlim, zeroline = F),
yaxis = list(range = ylim, zeroline = F),
shapes = shapes)
print(p)
}